Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
 -glucosyltransferase from
-glucosyltransferase from 
Hiromoto, Takeshi; Honjo, Eijiro*; Tamada, Taro; Kuroki, Ryota; Noda, Hisanobu*; Kazuma, Kohei*; Suzuki, Masahiko*
no journal, ,
UDP-glucose: anthocyanidin 3- -glucosyltransferase from
-glucosyltransferase from  (
( 3GT-A) catalyzes the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin. The glucosylation of delphinidin at the 3-hydroxyl group has been proposed as an initial glucosylation step toward the biosynthesis of ternatins, which are blue anthocyanins found in the petals of
3GT-A) catalyzes the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin. The glucosylation of delphinidin at the 3-hydroxyl group has been proposed as an initial glucosylation step toward the biosynthesis of ternatins, which are blue anthocyanins found in the petals of  . Although the crystal structures of several flavonoid glycosyltransferases (UGTs) were determined, the acceptor-substrate complexes were limited to the flavonol-bound forms. Here, in order to understand the acceptor-recognition scheme of
. Although the crystal structures of several flavonoid glycosyltransferases (UGTs) were determined, the acceptor-substrate complexes were limited to the flavonol-bound forms. Here, in order to understand the acceptor-recognition scheme of  3GT-A, the crystal structures in complex with anthocyanidin delphinidin and petunidin, and flavonol kaempferol were determined to resolutions of 2.6
3GT-A, the crystal structures in complex with anthocyanidin delphinidin and petunidin, and flavonol kaempferol were determined to resolutions of 2.6  , 2.7
, 2.7  , and 1.8
, and 1.8  respectively. The enzyme recognition of unstable anthocyanidins was firstly observed in this structural determination; nevertheless, the molecular orientations of these three acceptors in the binding site are different from those of the known flavonoid UGTs,
respectively. The enzyme recognition of unstable anthocyanidins was firstly observed in this structural determination; nevertheless, the molecular orientations of these three acceptors in the binding site are different from those of the known flavonoid UGTs, 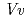 GT1 and UGT78G1. The crystal structures of
GT1 and UGT78G1. The crystal structures of  3GT-A provide insight not only into anthocyanidin configurations in enzyme, but also into a different binding scheme for acceptor-substrate recognition compared with the known UGTs.
3GT-A provide insight not only into anthocyanidin configurations in enzyme, but also into a different binding scheme for acceptor-substrate recognition compared with the known UGTs.
 -lactamase from moderate halophile
-lactamase from moderate halophileArai, Shigeki; Adachi, Motoyasu; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
A high content of acidic residues of halophilic proteins may destabilize the structure of halophilic protein and take away an enzymatic function under low salt concentration. However, periplasmic proteins of moderate halophiles require adapting a wide range of salt concentration (0.5 M - saturated NaCl). In this study, we investigated the structural and functional characteristics of HaBLA derived from periplasm of a moderate halophile  sp.560. By isothermal titration calorimetry, it was clarified that HaBLA hydrolyses penicillin G under 0 - 4 M NaCl. Moreover, by X-ray crystallographic analysis, we found the positive charges and the hydrophobic cluster near the entrance of the active site of HaBLA. These structural characteristics may attract penicillin G to the active site of HaBLA, which may participate in the low salt concentration adaptation of HaBLA.
sp.560. By isothermal titration calorimetry, it was clarified that HaBLA hydrolyses penicillin G under 0 - 4 M NaCl. Moreover, by X-ray crystallographic analysis, we found the positive charges and the hydrophobic cluster near the entrance of the active site of HaBLA. These structural characteristics may attract penicillin G to the active site of HaBLA, which may participate in the low salt concentration adaptation of HaBLA.
Kurihara, Kazuo; Tomoyori, Katsuaki; Tamada, Taro; Kuroki, Ryota
no journal, ,
Many of membrane proteins and protein complexes have larger molecular weight and then unit cells of their crystals have larger volume. Therefore, our group had designed the diffractometer which is able to cover such a crystal with large unit cell volume (target lattice length: 250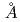 ). This proposal was accepted by Neutron Instrument Program Review Committee of J-PARC in September 2012. Larger unit cell volume causes a problem to separate spots closer to each other in spatial and time distribution in diffraction images. Therefore, our proposed diffractometer adoptted longer camera distance (800mm) and decoupled moderator as neutron source which has shorter pulse width. The neutron guide tube was designed to use limited surface of the decoupled moderator with high luminescence (40mm high
). This proposal was accepted by Neutron Instrument Program Review Committee of J-PARC in September 2012. Larger unit cell volume causes a problem to separate spots closer to each other in spatial and time distribution in diffraction images. Therefore, our proposed diffractometer adoptted longer camera distance (800mm) and decoupled moderator as neutron source which has shorter pulse width. The neutron guide tube was designed to use limited surface of the decoupled moderator with high luminescence (40mm high  60mm wide) whose luminosity is 1.24 times as high as that of average of the whole surface in the range over 2.86
60mm wide) whose luminosity is 1.24 times as high as that of average of the whole surface in the range over 2.86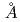 of wavelength. In addition, ellipsoidal shape in vertical design of the guide was adopted to suppress the number of neutron reflections at the guide mirror accompanying reduction of neutron intensity. In the horizontal design, curved shape was in part introduced to eliminate unnecessary
of wavelength. In addition, ellipsoidal shape in vertical design of the guide was adopted to suppress the number of neutron reflections at the guide mirror accompanying reduction of neutron intensity. In the horizontal design, curved shape was in part introduced to eliminate unnecessary  -rays and short-wavelength neutrons. According to ray-tracing simulation by McStas code, neutron flux at the sample position is estimated to be 5
-rays and short-wavelength neutrons. According to ray-tracing simulation by McStas code, neutron flux at the sample position is estimated to be 5 10
10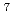 /cm
/cm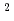 /s in the wavelength range of 1.5
/s in the wavelength range of 1.5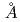 - 5.6
- 5.6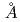 (first frame).
(first frame).
Tomoyori, Katsuaki; Kurihara, Kazuo; Tamada, Taro; Kuroki, Ryota
no journal, ,
In protein crystallography, the large biomacromolecules like the membrane proteins and protein complex are one of the most important research objects. It brings greatly contributions to the life science research field to elucidate the function by means of the structural analyses including hydrogen atom and hydration of these proteins. We propose to construct a new instrument to realize the neutron diffraction experiment using protein crystals with large unit cells. In scattering experiment with time-of-flight method, the most advantageous feature is to be able to improve the energy or time resolution by increasing flight path length. The long flight path can facilitate the spot separation from the measurement of protein crystals with large unit cells in the new instrument to be constructed in J-PARC/MLF. At present, we are predominantly planning to choose the decoupled moderator (DM) with comparatively narrow pulse width considering limited space to accommodate our new instrument inside J-PARC/MLF buildings. We evaluated the instrumental parameters to realize the measurement of protein crystal with large unit cell over 250  . The uncertainty of d-spacings is very large in low angle bank detector. It could be required to improve the angular divergence with some collimator and slit apparatus. We evaluated the capability of spot separation against scattering angles and determined the flight path length L1, L2 and the required angular divergence.
. The uncertainty of d-spacings is very large in low angle bank detector. It could be required to improve the angular divergence with some collimator and slit apparatus. We evaluated the capability of spot separation against scattering angles and determined the flight path length L1, L2 and the required angular divergence.
Tamada, Taro
no journal, ,
Most electron transfer proteins have cofactor molecules, such as heme, flavin, and iron-sulfur cluster, and transfer electrons via those cofactors. Therefore, it is important to obtain precise structure information including hydrogen atoms for understanding the electron transfer reaction. We have performed high-resolution neutron crystal structure analyses of two electron transfer proteins, high-potential iron-sulfur protein (HiPIP) and NADH-cytochrome 
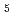 reductase (b5R). We succeeded in data collection of these proteins at higher resolution, 1.1
reductase (b5R). We succeeded in data collection of these proteins at higher resolution, 1.1  (HiPIP) and 1.4
(HiPIP) and 1.4  (b5R), using BL03 (iBIX) beamline in J-PARC/MLF. Joint neutron and X-ray crystallographic refinement is in progress, but we have already observed protonation statuses of polar residues located in molecular surface and orientation of water molecules. Furthermore, we confirmed some characteristic hydorogens which have unideal geometries. In this presentation, we also talk about our plan of new diffractomer in J-PARC, which is able to cover such a crystal with large unit cell (
(b5R), using BL03 (iBIX) beamline in J-PARC/MLF. Joint neutron and X-ray crystallographic refinement is in progress, but we have already observed protonation statuses of polar residues located in molecular surface and orientation of water molecules. Furthermore, we confirmed some characteristic hydorogens which have unideal geometries. In this presentation, we also talk about our plan of new diffractomer in J-PARC, which is able to cover such a crystal with large unit cell ( 250
250  ).
).
Hirayama, Hiroshi; Adachi, Motoyasu; Shimizu, Rumi; Kuroki, Ryota
no journal, ,
no abstracts in English
Yonetani, Yoshiteru
no journal, ,
no abstracts in English
Shimizu, Rumi; Hiromoto, Takeshi; Adachi, Motoyasu; Shibazaki, Chie; Kuroki, Ryota
no journal, ,
no abstracts in English
Hirano, Yu; Tamada, Taro; Kurihara, Kazuo; Kusaka, Katsuhiro*; Miki, Kunio*
no journal, ,
Information about hydrogen atoms are important for understanding both structure and function of proteins. The structure of hydrogen atoms in proteins has been discussed based on the ideal values of bond lengths and angles obtained from structures of small molecules. A huge contribution to the protein science will be made by the structure information of hydrogen atoms without restraints of the ideal values. Hydrogen atoms are easily detected in neutron crystal structures because neutrons have a strong diffraction power compared to other atoms in proteins. In this study, we have determined high-resolution neutron structure of the high-potential iron-sulfur protein (HiPIP). The neutron diffraction experiment was performed at the BL03 (iBIX) beamline of J-PARC/MLF. We have obtained the diffraction data set at 1.1 -resolution that is the highest resolution data in protein neutron studies. The protonation states of amino acid residues at the surface of HiPIP have been observed after structure refinement. In addition, many deviations from the ideal values were observed in the distances and angles involved in hydrogen atoms.
-resolution that is the highest resolution data in protein neutron studies. The protonation states of amino acid residues at the surface of HiPIP have been observed after structure refinement. In addition, many deviations from the ideal values were observed in the distances and angles involved in hydrogen atoms.
Sunami, Tomoko; Kono, Hidetoshi
no journal, ,
no abstracts in English
Nakagawa, Hiroshi
no journal, ,
Bio-molecular dynamics should be characterized with wide time-space ranges. Inelastic neutron scattering is the powerful method to observe the dynamics in THz region, which is essential for the solvent-coupled dynamics, and domain motions. Furthermore, this method is sensitive to the hydration water dynamics. The molecular dynamics simulation is effective for analysis and interpretation of the experimental data. We will show that the relationship between protein dynamical transition and hydration water dynamics, protein diffusion coupled with solvent and domain motions.
Uemura, Takuya*; Kita, Akiko*; Adachi, Motoyasu; Kuroki, Ryota; Morimoto, Yukio*
no journal, ,
no abstracts in English